Modeling mortality¶
Introduction¶
To design and evaluate an index based insurance contract, we need to understand the relationship between the index and losses incurred. Here we develop such a relationship using the z-scored NDVI to predict livestock mortality, using survey data from Marsabit.
Data¶
Mortality¶
Mortality was computed from survey data collected in Marsabit between 2008 and 2015. First read the data.
mort <- agrodata::data_ibli("marsabit_mortality.rds")
mort[1:5, 1:7]
## hhid season year sublocation type stock loss
## 1 1002 SRSD 2008 DAKABARICHA Camel 0.000000 0
## 2 1002 SRSD 2008 DAKABARICHA Cattle 13.000000 0
## 3 1002 SRSD 2008 DAKABARICHA Shoat 4.000000 0
## 4 1002 SRSD 2008 DAKABARICHA TLU 13.400000 0
## 5 1002 SRSD 2008 DAKABARICHA TLU per capita 1.914286 0
Load data with causes of death.
cause <- agrodata::data_ibli("marsabit_losses")
cause <- cause[,c("hhid", "sublocation", "cause","month", "year")]
colnames(cause)[4] <- "season"
s <- cause$season
cause$season <- "SRSD"
cause$season[s >=3 & s <=9] <-"LRLD"
cause <- na.omit(cause)
head(cause, n=2)
## hhid sublocation cause season year
## 342 1002 DAKABARICHA Starvation/Drought LRLD 2009
## 343 1002 DAKABARICHA Starvation/Drought LRLD 2009
Merge this table with mortality table and extract mortality causes by starvation/drought.
mort <- merge(mort, cause, by=c("hhid","sublocation", "season", "year"))
mort <- mort[mort$cause == "Starvation/Drought", ]
We mainly care about the mortality rates. Compute aggregates by year, season and sublocation.
mort <- mort[mort$type == "TLU", ]
amort <- aggregate(mort[,c("stock_beginning", "loss")], mort[ , c("year", "season", "sublocation")], sum, na.rm=TRUE)
head(amort, n=2)
## year season sublocation stock_beginning loss
## 1 2009 LRLD BUBISA 8936.4 2072.0
## 2 2010 LRLD BUBISA 520.3 40.4
amort$mortality_rate = 1 - ((amort$stock_beginning - amort$loss) / amort$stock_beginning )
amort$mortality_rate[amort$mortality_rate==0] <- NA
NDVI¶
Load the zNDVI data for 2000–2015 and income data for Marsabit that we computed in the previous section. We reshape the data from “wide” to “long” format.
LR <- agrodata::data_ibli("marsabit_zLRLD")
SR <- agrodata::data_ibli("marsabit_zSRSD")
LR[1:3, 1:6]
## sublocation LRLD2000 LRLD2001 LRLD2002 LRLD2003 LRLD2004
## 1 ARAPAL -2.0961337 0.3242631 -0.3526237 1.1115084 0.3370314
## 2 BADASA -0.6657791 0.2393530 0.6495006 0.9258476 0.6766376
## 3 BALESA -1.0856171 0.1699284 -0.2237794 1.4356138 -0.3569806
lr <- reshape(LR, direction="long", varying = 2:ncol(LR), v.names="NDVI", timevar="year", times=2000:2015)
lr$id <- "LRLD"
sr <- reshape(SR, direction="long", varying = 2:ncol(LR), v.names="NDVI", timevar="year", times=2000:2015)
sr$id <- "SRSD"
ndvi <- rbind(lr, sr)
colnames(ndvi)[4] <- "season"
ndvi$year_season <- apply(ndvi[, c("year", "season")], 1, function(x) paste(x, collapse="_"))
head(ndvi, n=2)
## sublocation year NDVI season year_season
## 1.2000 ARAPAL 2000 -2.0961337 LRLD 2000_LRLD
## 2.2000 BADASA 2000 -0.6657791 LRLD 2000_LRLD
Merge mortality and NDVI¶
d <- merge(ndvi, amort, by=c("year", "season", "sublocation"))
Explore the relation between mortality and NDVI.
cols <- c("red", "blue")
seas <- c("SRSD", "LRLD")
pcols <- cols[match(d$season, seas)]
plot(mortality_rate~NDVI, data=d, xlab="zndvi", ylab="mortality", col=pcols, pch=20)
legend("topright", seas, col=cols, pch=20)
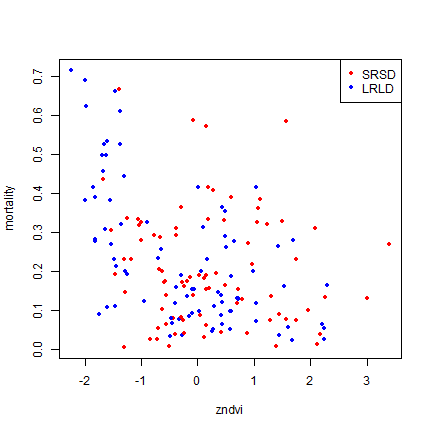
The relationship is a bit noisy (as to be expected). There is a lot of variability at zNDVI < -1. That could be a problem; as predictions of mortality at low zNDVI would be quite uncertain.
Mortality model¶
Build a local regression model.
dd <- na.omit(d[, c("NDVI", "mortality_rate")])
# add two "anchering points"
dd <- rbind(dd, c(-2.5, 0.9))
dd <- rbind(dd, c(-4, 1))
saveRDS(dd, "ndvi_mort.rds")
library(msir)
## Warning: package 'msir' was built under R version 4.3.1
## Package 'msir' version 1.3.3
## Type 'citation("msir")' for citing this R package in publications.
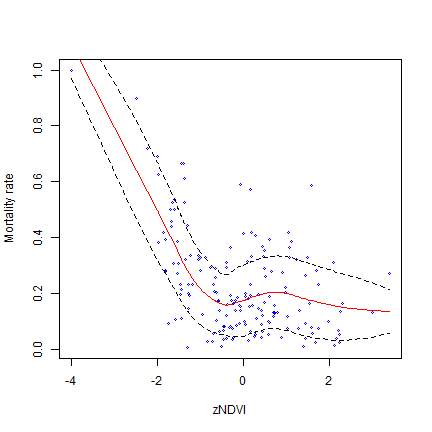
# Calculate and plot a 1.96 * SD prediction band
# that is a 95% prediction band
m <- loess.sd(dd)
plot(dd, cex=.5, col="blue", xlab="zNDVI", ylab="Mortality rate")
lines(m$x, m$y, col="red")
lines(m$x, m$upper, lty=2)
lines(m$x, m$lower, lty=2)
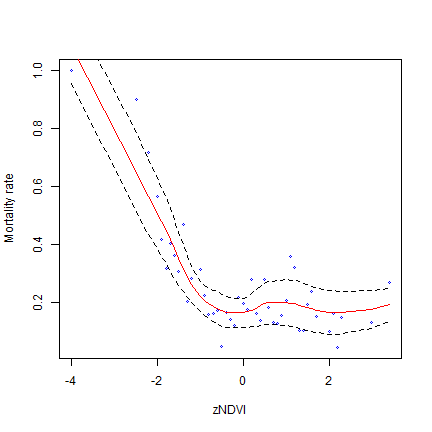
Same thing after a little generalization.
x <- dd
x$NDVI = round(x$NDVI, 1)
a <- aggregate(x[, "mortality_rate", drop=FALSE], x[, "NDVI", drop=FALSE], mean)
ma <- loess.sd(a)
plot(a, cex=.5, col="blue", xlab="zNDVI", ylab="Mortality rate")
lines(ma$x, ma$y, col="red")
lines(ma$x, ma$upper, lty=2)
lines(ma$x, ma$lower, lty=2)
Compare predicted with observed mortality rates
ml <- loess(mortality_rate ~ NDVI, data=dd)
p <- predict(ml, d)
plot(d$mortality_rate, p, xlab="mortality rate", ylab="predicted")
abline(0,1)
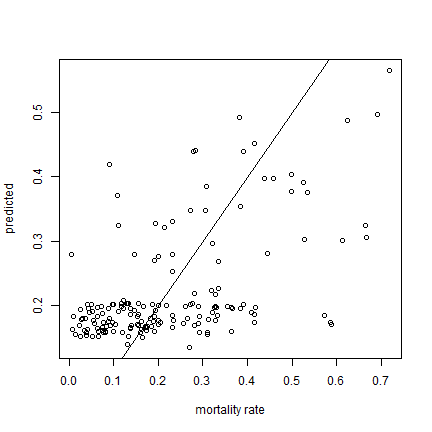
x <- na.omit(cbind(d$mortality_rate, p))
cor(x[,1], x[,2])
## [1] 0.5567935
d$predicted_mortality <- p
saveRDS(d, "pred_mort1.rds")
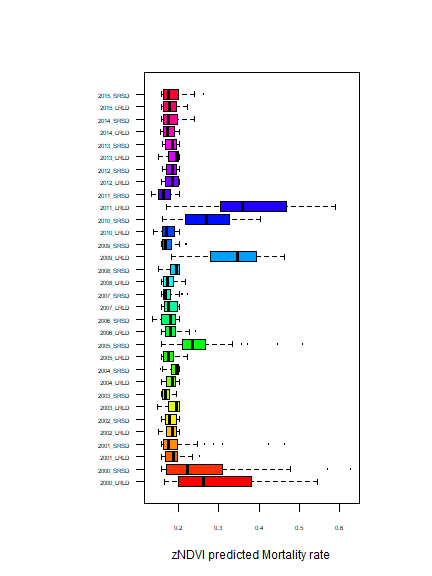
Predict mortality in other years based on their zNDVI.
ndvi$predicted_mortality <- predict(ml, ndvi)
par(mai=c(1,2,1,1))
boxplot(ndvi$predicted_mortality ~ ndvi$year_season, las=1, cex.axis=.5, horizontal=TRUE, xlab="zNDVI predicted Mortality rate", ylab="", cex=.25, col=rainbow(32))
saveRDS(ndvi, "pred_mort2.rds")